Initial Remark: Reload this page if formulas don’t display well!
As promised, here is the second part on how to obtain confidence intervals for fitted values obtained from nonlinear regression via nls or nlsLM (package ‘minpack.lm’).
I covered a Monte Carlo approach in https://rmazing.wordpress.com/2013/08/14/predictnls-part-1-monte-carlo-simulation-confidence-intervals-for-nls-models/, but here we will take a different approach: First- and second-order Taylor approximation around 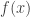 :
: 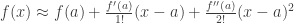 .
.
Using Taylor approximation for calculating confidence intervals is a matter of propagating the uncertainties of the parameter estimates obtained from vcov(model) to the fitted value. When using first-order Taylor approximation, this is also known as the “Delta method”. Those familiar with error propagation will know the formula
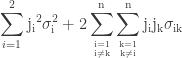 .
.
Heavily underused is the matrix notation of the famous formula above, for which a good derivation can be found at http://www.nada.kth.se/~kai-a/papers/arrasTR-9801-R3.pdf:
 ,
,
where 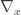 is the gradient vector of first-order partial derivatives and
is the gradient vector of first-order partial derivatives and 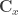 is the variance-covariance matrix. This formula corresponds to the first-order Taylor approximation. Now the problem with first-order approximations is that they assume linearity around
is the variance-covariance matrix. This formula corresponds to the first-order Taylor approximation. Now the problem with first-order approximations is that they assume linearity around 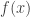 . Using the “Delta method” for nonlinear confidence intervals in R has been discussed in http://thebiobucket.blogspot.de/2011/04/fit-sigmoid-curve-with-confidence.html or http://finzi.psych.upenn.edu/R/Rhelp02a/archive/42932.html.
. Using the “Delta method” for nonlinear confidence intervals in R has been discussed in http://thebiobucket.blogspot.de/2011/04/fit-sigmoid-curve-with-confidence.html or http://finzi.psych.upenn.edu/R/Rhelp02a/archive/42932.html.
For highly nonlinear functions we need (at least) a second-order polynomial around 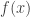 to realistically estimate the surrounding interval (red is linear approximation, blue is second-order polynomial on a sine function around
to realistically estimate the surrounding interval (red is linear approximation, blue is second-order polynomial on a sine function around  ):
):

Interestingly, there are also matrix-like notations for the second-order mean and variance in the literature (see http://dml.cz/dmlcz/141418 or http://iopscience.iop.org/0026-1394/44/3/012/pdf/0026-1394_44_3_012.pdf):
Second-order mean: ![\rm{E}[y] = f(\bar{x}_i) + \frac{1}{2}\rm{tr}(\mathbf{H}_{xx}\mathbf{C}_x)](https://s0.wp.com/latex.php?latex=%5Crm%7BE%7D%5By%5D+%3D+f%28%5Cbar%7Bx%7D_i%29+%2B+%5Cfrac%7B1%7D%7B2%7D%5Crm%7Btr%7D%28%5Cmathbf%7BH%7D_%7Bxx%7D%5Cmathbf%7BC%7D_x%29&bg=ffffff&fg=333333&s=0&c=20201002) .
.
Second-order variance: 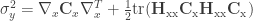 ,
,
where  is the Hessian matrix of second-order partial derivatives and
is the Hessian matrix of second-order partial derivatives and 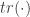 is the matrix trace (sum of diagonals).
is the matrix trace (sum of diagonals).
Enough theory, for wrapping this all up we need three utility functions:
1) numGrad for calculating numerical first-order partial derivatives.
numGrad <- function(expr, envir = .GlobalEnv)
{
f0 <- eval(expr, envir)
vars <- all.vars(expr)
p <- length(vars)
x <- sapply(vars, function(a) get(a, envir))
eps <- 1e-04
d <- 0.1
r <- 4
v <- 2
zero.tol <- sqrt(.Machine$double.eps/7e-07)
h0 <- abs(d * x) + eps * (abs(x) < zero.tol)
D <- matrix(0, length(f0), p)
Daprox <- matrix(0, length(f0), r)
for (i in 1:p) {
h <- h0
for (k in 1:r) {
x1 <- x2 <- x
x1 <- x1 + (i == (1:p)) * h
f1 <- eval(expr, as.list(x1))
x2 <- x2 - (i == (1:p)) * h
f2 <- eval(expr, envir = as.list(x2))
Daprox[, k] <- (f1 - f2)/(2 * h[i])
h <- h/v
}
for (m in 1:(r - 1)) for (k in 1:(r - m)) {
Daprox[, k] <- (Daprox[, k + 1] * (4^m) - Daprox[, k])/(4^m - 1)
}
D[, i] <- Daprox[, 1]
}
return(D)
}
2) numHess for calculating numerical second-order partial derivatives.
numHess <- function(expr, envir = .GlobalEnv)
{
f0 <- eval(expr, envir)
vars <- all.vars(expr)
p <- length(vars)
x <- sapply(vars, function(a) get(a, envir))
eps <- 1e-04
d <- 0.1
r <- 4
v <- 2
zero.tol <- sqrt(.Machine$double.eps/7e-07)
h0 <- abs(d * x) + eps * (abs(x) < zero.tol)
Daprox <- matrix(0, length(f0), r)
Hdiag <- matrix(0, length(f0), p)
Haprox <- matrix(0, length(f0), r)
H <- matrix(NA, p, p)
for (i in 1:p) {
h <- h0
for (k in 1:r) {
x1 <- x2 <- x
x1 <- x1 + (i == (1:p)) * h
f1 <- eval(expr, as.list(x1))
x2 <- x2 - (i == (1:p)) * h
f2 <- eval(expr, envir = as.list(x2))
Haprox[, k] <- (f1 - 2 * f0 + f2)/h[i]^2
h <- h/v
}
for (m in 1:(r - 1)) for (k in 1:(r - m)) {
Haprox[, k] <- (Haprox[, k + 1] * (4^m) - Haprox[, k])/(4^m - 1)
}
Hdiag[, i] <- Haprox[, 1]
}
for (i in 1:p) {
for (j in 1:i) {
if (i == j) {
H[i, j] <- Hdiag[, i]
}
else {
h <- h0
for (k in 1:r) {
x1 <- x2 <- x
x1 <- x1 + (i == (1:p)) * h + (j == (1:p)) *
h
f1 <- eval(expr, as.list(x1))
x2 <- x2 - (i == (1:p)) * h - (j == (1:p)) *
h
f2 <- eval(expr, envir = as.list(x2))
Daprox[, k] <- (f1 - 2 * f0 + f2 - Hdiag[, i] * h[i]^2 - Hdiag[, j] * h[j]^2)/(2 * h[i] * h[j])
h <- h/v
}
for (m in 1:(r - 1)) for (k in 1:(r - m)) {
Daprox[, k] <- (Daprox[, k + 1] * (4^m) - Daprox[, k])/(4^m - 1)
}
H[i, j] <- H[j, i] <- Daprox[, 1]
}
}
}
return(H)
}
And a small function for the matrix trace:
tr <- function(mat) sum(diag(mat), na.rm = TRUE)
1) and 2) are modified versions of the genD function in the “numDeriv” package that can handle expressions.
Now we need the predictNLS function that wraps it all up:
predictNLS <- function(
object,
newdata,
interval = c("none", "confidence", "prediction"),
level = 0.95,
...
)
{
require(MASS, quietly = TRUE)
interval <- match.arg(interval)
## get right-hand side of formula
RHS <- as.list(object$call$formula)[[3]]
EXPR <- as.expression(RHS)
## all variables in model
VARS <- all.vars(EXPR)
## coefficients
COEF <- coef(object)
## extract predictor variable
predNAME <- setdiff(VARS, names(COEF))
## take fitted values, if 'newdata' is missing
if (missing(newdata)) {
newdata <- eval(object$data)[predNAME]
colnames(newdata) <- predNAME
}
## check that 'newdata' has same name as predVAR
if (names(newdata)[1] != predNAME) stop("newdata should have name '", predNAME, "'!")
## get parameter coefficients
COEF <- coef(object)
## get variance-covariance matrix
VCOV <- vcov(object)
## augment variance-covariance matrix for 'mvrnorm'
## by adding a column/row for 'error in x'
NCOL <- ncol(VCOV)
ADD1 <- c(rep(0, NCOL))
ADD1 <- matrix(ADD1, ncol = 1)
colnames(ADD1) <- predNAME
VCOV <- cbind(VCOV, ADD1)
ADD2 <- c(rep(0, NCOL + 1))
ADD2 <- matrix(ADD2, nrow = 1)
rownames(ADD2) <- predNAME
VCOV <- rbind(VCOV, ADD2)
NR <- nrow(newdata)
respVEC <- numeric(NR)
seVEC <- numeric(NR)
varPLACE <- ncol(VCOV)
outMAT <- NULL
## define counter function
counter <- function (i)
{
if (i%%10 == 0)
cat(i)
else cat(".")
if (i%%50 == 0)
cat("\n")
flush.console()
}
## calculate residual variance
r <- residuals(object)
w <- weights(object)
rss <- sum(if (is.null(w)) r^2 else r^2 * w)
df <- df.residual(object)
res.var <- rss/df
## iterate over all entries in 'newdata' as in usual 'predict.' functions
for (i in 1:NR) {
counter(i)
## get predictor values and optional errors
predVAL <- newdata[i, 1]
if (ncol(newdata) == 2) predERROR <- newdata[i, 2] else predERROR <- 0
names(predVAL) <- predNAME
names(predERROR) <- predNAME
## create mean vector
meanVAL <- c(COEF, predVAL)
## create augmented variance-covariance matrix
## by putting error^2 in lower-right position of VCOV
newVCOV <- VCOV
newVCOV[varPLACE, varPLACE] <- predERROR^2
SIGMA <- newVCOV
## first-order mean: eval(EXPR), first-order variance: G.S.t(G)
MEAN1 <- try(eval(EXPR, envir = as.list(meanVAL)), silent = TRUE)
if (inherits(MEAN1, "try-error")) stop("There was an error in evaluating the first-order mean!")
GRAD <- try(numGrad(EXPR, as.list(meanVAL)), silent = TRUE)
if (inherits(GRAD, "try-error")) stop("There was an error in creating the numeric gradient!")
VAR1 <- GRAD %*% SIGMA %*% matrix(GRAD)
## second-order mean: firstMEAN + 0.5 * tr(H.S),
## second-order variance: firstVAR + 0.5 * tr(H.S.H.S)
HESS <- try(numHess(EXPR, as.list(meanVAL)), silent = TRUE)
if (inherits(HESS, "try-error")) stop("There was an error in creating the numeric Hessian!")
valMEAN2 <- 0.5 * tr(HESS %*% SIGMA)
valVAR2 <- 0.5 * tr(HESS %*% SIGMA %*% HESS %*% SIGMA)
MEAN2 <- MEAN1 + valMEAN2
VAR2 <- VAR1 + valVAR2
## confidence or prediction interval
if (interval != "none") {
tfrac <- abs(qt((1 - level)/2, df))
INTERVAL <- tfrac * switch(interval, confidence = sqrt(VAR2),
prediction = sqrt(VAR2 + res.var))
LOWER <- MEAN2 - INTERVAL
UPPER <- MEAN2 + INTERVAL
names(LOWER) <- paste((1 - level)/2 * 100, "%", sep = "")
names(UPPER) <- paste((1 - (1- level)/2) * 100, "%", sep = "")
} else {
LOWER <- NULL
UPPER <- NULL
}
RES <- c(mu.1 = MEAN1, mu.2 = MEAN2, sd.1 = sqrt(VAR1), sd.2 = sqrt(VAR2), LOWER, UPPER)
outMAT <- rbind(outMAT, RES)
}
cat("\n")
rownames(outMAT) <- NULL
return(outMAT)
}
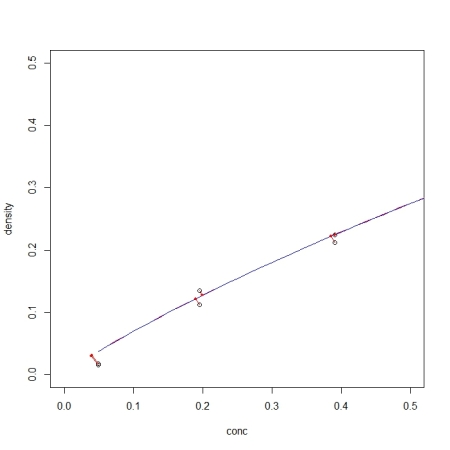
With all functions at hand, we can now got through the same example as used in the Monte Carlo post:
DNase1 <- subset(DNase, Run == 1)
fm1DNase1 <- nls(density ~ SSlogis(log(conc), Asym, xmid, scal), DNase1)
> predictNLS(fm1DNase1, newdata = data.frame(conc = 5), interval = "confidence")
.
mu.1 mu.2 sd.1 sd.2 2.5% 97.5%
[1,] 1.243631 1.243288 0.03620415 0.03620833 1.165064 1.321511
The errors/confidence intervals are larger than with the MC approch (who knows why?) but it is very interesting to see how close the second-order corrected mean (1.243288) comes to the mean of the simulated values from the Monte Carlo approach (1.243293)!
The two approach (MC/Taylor) will be found in the predictNLS function that will be part of the “propagate” package in a few days at CRAN…
Cheers,
Andrej
using
(1), where
is the gradient and
the covariance matrix of the coefficients
, is called the “Delta Method” and is widely applied in nonlinear least-squares (NLS) fitting. However, this method is based on first-order Taylor expansion and thus assummes linearity around
. The second-order approach can partially correct for this restriction by using a second-order polynomial around
, which is
(2), where
is the matrix trace and
is the Hessian.
(3) or
(4), respectively, where the residual variance
(5).
in the prediction interval can be implemented as an extended gradient and “augmented” covariance matrix. So instead of using
(6), we take
(7) as the expression and augment the
covariance matrix
to an
covariance matrix, where
. Partial differentiation and matrix multiplication will then yield, for example with two coefficients
and
and their corresponding covariance matrix
:
(8)
(9)
, which then goes into (4).
. All
simulations are then evaluated with (7) and the usual
quantiles calculated for the prediction interval. Using the original covariance matrix with (6) will deliver the MC-based confidence interval.
are quite asymmetric, which the classical Delta Method does not capture:

 Posted by anspiess
Posted by anspiess